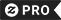Tap / click on image to see more RealViewsTM
£31.10
per shirt
im not lazy i just dont have enough enzymes T-Shirt
Qty:
Style
Womens Basic T-Shirt
Colour & Print Process
White
Classic Printing: No Underbase
Vivid Printing: White Underbase
+£7.70
+£7.70
+£7.70
About T-Shirts
Sold by
About This Design
im not lazy i just dont have enough enzymes T-Shirt
Enzymes are proteins that catalyse (i.e., increase the rates of) chemical reactions.[1][2] In enzymatic reactions, the molecules at the beginning of the process are called substrates, and the enzyme converts them into different molecules, called the products. Almost all processes in a biological cell need enzymes to occur at significant rates. Since enzymes are selective for their substrates and speed up only a few reactions from among many possibilities, the set of enzymes made in a cell determines which metabolic pathways occur in that cell. Like all catalysts, enzymes work by lowering the activation energy (Ea‡) for a reaction, thus dramatically increasing the rate of the reaction. As a result, products are formed faster and reactions reach their equilibrium state more rapidly. Most enzyme reaction rates are millions of times faster than those of comparable un-catalysed reactions. As with all catalysts, enzymes are not consumed by the reactions they catalyse, nor do they alter the equilibrium of these reactions. However, enzymes do differ from most other catalysts by being much more specific. Enzymes are known to catalyse about 4,000 biochemical reactions.[3] A few RNA molecules called ribozymes also catalyse reactions, with an important example being some parts of the ribosome.[4][5] Synthetic molecules called artificial enzymes also display enzyme-like catalysis.[6] Enzyme activity can be affected by other molecules. Inhibitors are molecules that decrease enzyme activity; activators are molecules that increase activity. Many drugs and poisons are enzyme inhibitors. Activity is also affected by temperature, chemical environment (e.g., pH), and the concentration of substrate. Some enzymes are used commercially, for example, in the synthesis of antibiotics. In addition, some household products use enzymes to speed up biochemical reactions (e.g., enzymes in biological washing powders break down protein or fat stains on clothes; enzymes in meat tenderizers break down proteins, making the meat easier to chew). Contents [hide] 1 Etymology and history 2 Structures and mechanisms 2.1 Specificity 2.1.1 "Lock and key" model 2.2 Mechanisms 2.2.1 Transition State Stabilisation 2.2.2 Dynamics and function 2.3 Allosteric modulation 3 Cofactors and coenzymes 3.1 Cofactors 3.2 Coenzymes 4 Thermodynamics 5 Kinetics 6 Inhibition 7 Biological function 8 Control of activity 9 Involvement in disease 10 Naming conventions 11 Industrial applications 12 See also 13 References 14 Further reading 15 External links [edit] Etymology and history Eduard BuchnerAs early as the late 18th and early 19th centuries, the digestion of meat by stomach secretions[7] and the conversion of starch to sugars by plant extracts and saliva were known. However, the mechanism by which this occurred had not been identified.[8] In the 19th century, when studying the fermentation of sugar to alcohol by yeast, Louis Pasteur came to the conclusion that this fermentation was catalysed by a vital force contained within the yeast cells called "ferments", which were thought to function only within living organisms. He wrote that "alcoholic fermentation is an act correlated with the life and organisation of the yeast cells, not with the death or putrefaction of the cells."[9] In 1877, German physiologist Wilhelm Kühne (1837–1900) first used the term enzyme, which comes from ενζυμον, "in leaven", to describe this process.[10] The word enzyme was used later to refer to nonliving substances such as pepsin, and the word ferment was used to refer to chemical activity produced by living organisms. In 1897, Eduard Buchner submitted his first paper on the ability of yeast extracts that lacked any living yeast cells to ferment sugar. In a series of experiments at the of Berlin, he found that the sugar was fermented even when there were no living yeast cells in the mixture.[11] He named the enzyme that brought about the fermentation of sucrose "zymase".[12] In 1907, he received the Nobel Prize in Chemistry "for his biochemical research and his discovery of cell-free fermentation". Following Buchner's example, enzymes are usually named according to the reaction they carry out. Typically, to generate the name of an enzyme, the suffix -ase is added to the name of its substrate (e.g., lactase is the enzyme that cleaves lactose) or the type of reaction (e.g., DNA polymerase forms DNA polymers).[13] Having shown that enzymes could function outside a living cell, the next step was to determine their biochemical nature. Many early workers noted that enzymatic activity was associated with proteins, but several scientists (such as Nobel laureate Richard Willstätter) argued that proteins were merely carriers for the true enzymes and that proteins per se were incapable of catalysis. However, in 1926, James B. Sumner showed that the enzyme urease was a pure protein and crystallised it; Sumner did likewise for the enzyme catalase in 1937. The conclusion that pure proteins can be enzymes was definitively proved by Northrop and Stanley, who worked on the digestive enzymes pepsin (1930), trypsin and chymotrypsin. These three scientists were awarded the 1946 Nobel Prize in Chemistry.[14] This discovery that enzymes could be crystallised eventually allowed their structures to be solved by x-ray crystallography. This was first done for lysozyme, an enzyme found in tears, saliva and egg whites that digests the coating of some bacteria; the structure was solved by a group led by David Chilton Phillips and published in 1965.[15] This high-resolution structure of lysozyme marked the beginning of the field of structural biology and the effort to understand how enzymes work at an atomic level of detail. [edit] Structures and mechanisms See also: Enzyme catalysis Ribbon diagram showing human carbonic anhydrase II. The grey sphere is the zinc cofactor in the active site. Diagram drawn from PDB 1MOO.Enzymes are generally globular proteins and range from just 62 amino acid residues in size, for the monomer of 4-oxalocrotonate tautomerase,[16] to over 2,500 residues in the animal fatty acid synthase.[17] A small number of RNA-based biological catalysts exist, with the most common being the ribosome; these are referred to as either RNA-enzymes or ribozymes. The activities of enzymes are determined by their three-dimensional structure.[18] However, although structure does determine function, predicting a novel enzyme's activity just from its structure is a very difficult problem that has not yet been solved.[19] Most enzymes are much larger than the substrates they act on, and only a small portion of the enzyme (around 3–4 amino acids) is directly involved in catalysis.[20] The region that contains these catalytic residues, binds the substrate, and then carries out the reaction is known as the active site. Enzymes can also contain sites that bind cofactors, which are needed for catalysis. Some enzymes also have binding sites for small molecules, which are often direct or indirect products or substrates of the reaction catalysed. This binding can serve to increase or decrease the enzyme's activity, providing a means for feedback regulation. Like all proteins, enzymes are long, linear chains of amino acids that fold to produce a three-dimensional product. Each unique amino acid sequence produces a specific structure, which has unique properties. Individual protein chains may sometimes group together to form a protein complex. Most enzymes can be denatured—that is, unfolded and inactivated—by heating or chemical denaturants, which disrupt the three-dimensional structure of the protein. Depending on the enzyme, denaturation may be reversible or irreversible. Structures of enzymes in complex with substrates or substrate analogs during a reaction may be obtained using Time resolved crystallography methods. [edit] Specificity Enzymes are usually very specific as to which reactions they catalyse and the substrates that are involved in these reactions. Complementary shape, charge and hydrophilic/hydrophobic characteristics of enzymes and substrates are responsible for this specificity. Enzymes can also show impressive levels of stereospecificity, regioselectivity and chemoselectivity.[21] Some of the enzymes showing the highest specificity and accuracy are involved in the copying and expression of the genome. These enzymes have "proof-reading" mechanisms. Here, an enzyme such as DNA polymerase catalyses a reaction in a first step and then checks that the product is correct in a second step.[22] This two-step process results in average error rates of less than 1 error in 100 million reactions in high-fidelity mammalian polymerases.[23] Similar proofreading mechanisms are also found in RNA polymerase,[24] aminoacyl tRNA synthetases[25] and ribosomes.[26] Some enzymes that produce secondary metabolites are described as promiscuous, as they can act on a relatively broad range of different substrates. It has been suggested that this broad substrate specificity is important for the evolution of new biosynthetic pathways.[27] [edit] "Lock and key" model Enzymes are very specific, and it was suggested by the Nobel laureate organic chemist Emil Fischer in 1894 that this was because both the enzyme and the substrate possess specific complementary geometric shapes that fit exactly into one another.[28] This is often referred to as "the lock and key" model. However, while this model explains enzyme specificity, it fails to explain the stabilisation of the transition state that enzymes achieve. Diagrams to show the induced fit hypothesis of enzyme action.In 1958, Daniel Koshland suggested a modification to the lock and key model: since enzymes are rather flexible structures, the active site is continually reshaped by interactions with the substrate as the substrate interacts with the enzyme.[29] As a result, the substrate does not simply bind to a rigid active site; the amino acid side chains which make up the active site are moulded into the precise positions that enable the enzyme to perform its catalytic function. In some cases, such as glycosidases, the substrate molecule also changes shape slightly as it enters the active site.[30] The active site continues to change until the substrate is completely bound, at which point the final shape and charge is determined.[31] Induced fit may enhance the fidelity of molecular recognition in the presence of competition and noise via the conformational proofreading mechanism .[32] [edit] Mechanisms Enzymes can act in several ways, all of which lower ΔG‡:[33] Lowering the activation energy by creating an environment in which the transition state is stabilised (e.g. straining the shape of a substrate—by binding the transition-state conformation of the substrate/product molecules, the enzyme distorts the bound substrate(s) into their transition state form, thereby reducing the amount of energy required to complete the transition). Lowering the energy of the transition state, but without distorting the substrate, by creating an environment with the opposite charge distribution to that of the transition state. Providing an alternative pathway. For example, temporarily reacting with the substrate to form an intermediate ES complex, which would be impossible in the absence of the enzyme. Reducing the reaction entropy change by bringing substrates together in the correct orientation to react. Considering ΔH‡ alone overlooks this effect. Increases in temperatures speed up reactions. Thus, temperature increases help the enzyme function and develop the end product even faster. However, if heated too much, the enzyme’s shape deteriorates and only when the temperature comes back to normal does the enzyme regain its shape. Some enzymes like thermolabile enzymes work best at low temperatures. Interestingly, this entropic effect involves destabilisation of the ground state,[34] and its contribution to catalysis is relatively small.[35] [edit] Transition State Stabilisation The understanding of the origin of the reduction of ΔG‡ requires one to find out how the enzymes can stabilise its transition state more than the transition state of the uncatalyzed reaction. Apparently, the most effective way for reaching large stabilisation is the use of electrostatic effects, in particular, by having a relatively fixed polar environment that is orientated towards the charge distribution of the transition state.[36] Such an environment does not exist in the uncatalyzed reaction in water. [edit] Dynamics and function See also: Protein dynamics The internal dynamics of enzymes is linked to their mechanism of catalysis.[37][38][39] Internal dynamics are the movement of parts of the enzyme's structure, such as individual amino acid residues, a group of amino acids, or even an entire protein domain. These movements occur at various time-scales ranging from femtoseconds to seconds. Networks of protein residues throughout an enzyme's structure can contribute to catalysis through dynamic motions.[40][41][42][43] Protein motions are vital to many enzymes, but whether small and fast vibrations, or larger and slower conformational movements are more important depends on the type of reaction involved. However, although these movements are important in binding and releasing substrates and products, it is not clear if protein movements help to accelerate the chemical steps in enzymatic reactions.[44] These new insights also have implications in understanding allosteric effects and developing new drugs. [edit] Allosteric modulation Allosteric transition of an enzyme between R and T states, stabilised by an agonist, an inhibitor and a substrate (the MWC model)Main article: Allosteric regulation Allosteric sites are sites on the enzyme that bind to molecules in the cellular environment. The sites form weak, noncovalent bonds with these molecules, causing a change in the conformation of the enzyme. This change in conformation translates to the active site, which then affects the reaction rate of the enzyme.[45] Allosteric interactions can both inhibit and activate enzymes and are a common way that enzymes are controlled in the body.[46] [edit] Cofactors and coenzymes Main articles: Cofactor (biochemistry) and Coenzyme [edit] Cofactors Some enzymes do not need any additional components to show full activity. However, others require non-protein molecules called cofactors to be bound for activity.[47] Cofactors can be either inorganic (e.g., metal ions and iron-sulphur clusters) or organic compounds (e.g., flavin and heme). Organic cofactors can be either prosthetic groups, which are tightly bound to an enzyme, or coenzymes, which are released from the enzyme's active site during the reaction. Coenzymes include NADH, NADPH and adenosine triphosphate. These molecules transfer chemical groups between enzymes.[48] An example of an enzyme that contains a cofactor is carbonic anhydrase, and is shown in the ribbon diagram above with a zinc cofactor bound as part of its active site.[49] These tightly bound molecules are usually found in the active site and are involved in catalysis. For example, flavin and heme cofactors are often involved in redox reactions. Enzymes that require a cofactor but do not have one bound are called apoenzymes or apoproteins. An apoenzyme together with its cofactor(s) is called a holoenzyme (this is the active form). Most cofactors are not covalently attached to an enzyme, but are very tightly bound. However, organic prosthetic groups can be covalently bound (e.g., thiamine pyrophosphate in the enzyme pyruvate dehydrogenase). The term "holoenzyme" can also be applied to enzymes that contain multiple protein subunits, such as the DNA polymerases; here the holoenzyme is the complete complex containing all the subunits needed for activity. [edit] Coenzymes Space-filling model of the coenzyme NADHCoenzymes are small organic molecules that transport chemical groups from one enzyme to another.[50] Some of these chemicals such as riboflavin, thiamine and folic acid are vitamins (compounds which cannot be synthesized by the body and must be acquired from the diet). The chemical groups carried include the hydride ion (H-) carried by NAD or NADP+, the phosphate group carried by adenosine triphosphate, the acetyl group carried by coenzyme A, formyl, methenyl or methyl groups carried by folic acid and the methyl group carried by S-adenosylmethionine. Since coenzymes are chemically changed as a consequence of enzyme action, it is useful to consider coenzymes to be a special class of substrates, or second substrates, which are common to many different enzymes. For example, about 700 enzymes are known to use the coenzyme NADH.[51] Coenzymes are usually continuously regenerated and their concentrations maintained at a steady level inside the cell: for example, NADPH is regenerated through the pentose phosphate pathway and S-adenosylmethionine by methionine adenosyltransferase. This continuous regeneration means that even small amounts of coenzymes are used very intensively. For example, the human body turns over its own weight in ATP each day.[52] [edit] Thermodynamics The energies of the stages of a chemical reaction. Substrates need a lot of energy to reach a transition state, which then decays into products. The enzyme stabilises the transition state, reducing the energy needed to form products.Main articles: Activation energy, Thermodynamic equilibrium, and Chemical equilibrium As all catalysts, enzymes do not alter the position of the chemical equilibrium of the reaction. Usually, in the presence of an enzyme, the reaction runs in the same direction as it would without the enzyme, just more quickly. However, in the absence of the enzyme, other possible uncatalyzed, "spontaneous" reactions might lead to different products, because in those conditions this different product is formed faster. Furthermore, enzymes can couple two or more reactions, so that a thermodynamically favourable reaction can be used to "drive" a thermodynamically unfavourable one. For example, the hydrolysis of ATP is often used to drive other chemical reactions.[53] Enzymes catalyse the forward and backward reactions equally. They do not alter the equilibrium itself, but only the speed at which it is reached. For example, carbonic anhydrase catalyses its reaction in either direction depending on the concentration of its reactants. (in tissues; high CO2 concentration) (in lungs; low CO2 concentration) Nevertheless, if the equilibrium is greatly displaced in one direction, that is, in a very exergonic reaction, the reaction is effectively irreversible. Under these conditions the enzyme will, in fact, only catalyse the reaction in the thermodynamically allowed direction. [edit] Kinetics Main article: Enzyme kinetics Mechanism for a single substrate enzyme catalysed reaction. The enzyme (E) binds a substrate (S) and produces a product (P).Enzyme kinetics is the investigation of how enzymes bind substrates and turn them into products. The rate data used in kinetic analyses are obtained from enzyme assays. In 1902 Victor Henri[54] proposed a quantitative theory of enzyme kinetics, but his experimental data were not useful because the significance of the hydrogen ion concentration was not yet appreciated. After Peter Lauritz Sørensen had defined the logarithmic pH-scale and introduced the concept of buffering in 1909[55] the German chemist Leonor Michaelis and his Canadian postdoc Maud Leonora Menten repeated Henri's experiments and confirmed his equation which is referred to as Henri-Michaelis-Menten kinetics (sometimes also Michaelis-Menten kinetics).[56] Their work was further developed by G. E. Briggs and J. B. S. Haldane, who derived kinetic equations that are still widely used today.[57] The major contribution of Henri was to think of enzyme reactions in two stages. In the first, the substrate binds reversibly to the enzyme, forming the enzyme-substrate complex. This is sometimes called the Michaelis complex. The enzyme then catalyses the chemical step in the reaction and releases the product. Saturation curve for an enzyme reaction showing the relation between the substrate concentration (S) and rate (v).Enzymes can catalyse up to several million reactions per second. For example, the uncatalyzed decarboxylation of orotidine 5'-monophosphate has a half life of 78 million years. However, when the enzyme orotidine 5'-phosphate decarboxylase is added, the same process takes just 25 milliseconds.[58] Enzyme rates depend on solution conditions and substrate concentration. Conditions that denature the protein abolish enzyme activity, such as high temperatures, extremes of pH or high salt concentrations, while raising substrate concentration tends to increase activity. To find the maximum speed of an enzymatic reaction, the substrate concentration is increased until a constant rate of product formation is seen. This is shown in the saturation curve on the right. Saturation happens because, as substrate concentration increases, more and more of the free enzyme is converted into the substrate-bound ES form. At the maximum velocity (Vmax) of the enzyme, all the enzyme active sites are bound to substrate, and the amount of ES complex is the same as the total amount of enzyme. However, Vmax is only one kinetic constant of enzymes. The amount of substrate needed to achieve a given rate of reaction is also important. This is given by the Michaelis-Menten constant (Km), which is the substrate concentration required for an enzyme to reach one-half its maximum velocity. Each enzyme has a characteristic Km for a given substrate, and this can show how tight the binding of the substrate is to the enzyme. Another useful constant is kcat, which is the number of substrate molecules handled by one active site per second. The efficiency of an enzyme can be expressed in terms of kcat/Km. This is also called the specificity constant and incorporates the rate constants for all steps in the reaction. Because the specificity constant reflects both affinity and catalytic ability, it is useful for comparing different enzymes against each other, or the same enzyme with different substrates. The theoretical maximum for the specificity constant is called the diffusion limit and is about 108 to 109 (M−1 s−1). At this point every collision of the enzyme with its substrate will result in catalysis, and the rate of product formation is not limited by the reaction rate but by the diffusion rate. Enzymes with this property are called catalytically perfect or kinetically perfect. Example of such enzymes are triose-phosphate isomerase, carbonic anhydrase, acetylcholinesterase, catalase, fumarase, β-lactamase, and superoxide dismutase. Michaelis-Menten kinetics relies on the law of mass action, which is derived from the assumptions of free diffusion and thermodynamically driven random collision. However, many biochemical or cellular processes deviate significantly from these conditions, because of macromolecular crowding, phase-separation of the enzyme/substrate/product, or one or two-dimensional molecular movement.[59] In these situations, a fractal Michaelis-Menten kinetics may be applied.[60][61][62][63] Some enzymes operate with kinetics which are faster than diffusion rates, which would seem to be impossible. Several mechanisms have been invoked to explain this phenomenon. Some proteins are believed to accelerate catalysis by drawing their substrate in and pre-orientating them by using dipolar electric fields. Other models invoke a quantum-mechanical tunnelling explanation, whereby a proton or an electron can tunnel through activation barriers, although for proton tunnelling this model remains somewhat controversial.[64][65] Quantum tunnelling for protons has been observed in tryptamine.[66] This suggests that enzyme catalysis may be more accurately chraacterised as "through the barrier" rather than the traditional model, which requires substrates to go "over" a lowered energy barrier. [edit] Inhibition Competitive inhibitors bind reversibly to the enzyme, preventing the binding of substrate. On the other hand, binding of substrate prevents binding of the inhibitor. Substrate and inhibitor compete for the enzyme. Types of inhibition. This classification was introduced by W.W. Cleland.[67]Main article: Enzyme inhibitor Enzyme reaction rates can be decreased by various types of enzyme inhibitors. Competitive inhibition In competitive inhibition, the inhibitor and substrate compete for the enzyme (i.e., they can not bind at the same time).[68] Often competitive inhibitors strongly resemble the real substrate of the enzyme. For example, methotrexate is a competitive inhibitor of the enzyme dihydrofolate reductase, which catalyses the reduction of dihydrofolate to tetrahydrofolate. The similarity between the structures of folic acid and this drug are shown in the figure to the right bottom. Note that binding of the inhibitor need not be to the substrate binding site (as frequently stated), if binding of the inhibitor changes the conformation of the enzyme to prevent substrate binding and vice versa. In competitive inhibition the maximal velocity of the reaction is not changed, but higher substrate concentrations are required to reach a given velocity, increasing the apparent Km. Uncompetitive inhibition In uncompetitive inhibition the inhibitor can not bind to the free enzyme, but only to the ES-complex. The EIS-complex thus formed is enzymatically inactive. This type of inhibition is rare, but may occur in multimeric enzymes. Non-competitive inhibition Non-competitive inhibitors can bind to the enzyme at the same time as the substrate, i.e. they never bind to the active site. Both the EI and EIS complexes are enzymatically inactive. Because the inhibitor can not be driven from the enzyme by higher substrate concentration (in contrast to competitive inhibition), the apparent Vmax changes. But because the substrate can still bind to the enzyme, the Km stays the same. Mixed inhibition This type of inhibition resembles the non-competitive, except that the EIS-complex has residual enzymatic activity. In many organisms inhibitors may act as part of a feedback mechanism. If an enzyme produces too much of one substance in the organism, that substance may act as an inhibitor for the enzyme at the beginning of the pathway that produces it, causing production of the substance to slow down or stop when there is sufficient amount. This is a form of negative feedback. Enzymes which are subject to this form of regulation are often multimeric and have allosteric binding sites for regulatory substances. Their substrate/velocity plots are not hyperbolar, but sigmoidal (S-shaped).
Store
Customer Reviews
4.6 out of 5 stars rating15.2K Total Reviews
15,198 Reviews
Reviews for similar products
5 out of 5 stars rating
By Tanya R.11 September 2022 • Verified Purchase
Womens Basic T-Shirt, White, Adult L
Zazzle Reviewer Program
When my teeshirt arrived I was delighted. The material is very good quality and it fitted me PERFECT.
I think ZAZZLE is much better than other companies who sell teeshirts.
I highly recommend anyone to shop here!
THANKU ZAZZLE! 😀😁🙌. Printing on my teeshirt was perfect.
5 out of 5 stars rating
By Scriptural G.5 July 2020 • Verified Purchase
Womens V-Neck T-Shirt, White, Adult S
Creator Review
I designed this shirt and I’m so happy with the print. I bought it in the cheaper shirt available and I bought it 2 sizes bigger because it runs small. It was perfect to wear to church today! It printed out beautifully! I’m so impressed with the quality!
5 out of 5 stars rating
By David A.10 June 2023 • Verified Purchase
Womens Basic T-Shirt, White, Adult M
Zazzle Reviewer Program
Respecful to the design. Beautiful, as expected from Zazzle
Tags
Other Info
Product ID: 235136738702665944
Created on 11/10/2010, 22:07
Rating: G
Recently Viewed Items